Command Line Usage
This section provides more “reference manual” style documentation when compared with Getting Started.
After installation, you can run clinvar-this:
$ clinvar-this --help
usage: clinvar-this [-h] [--verbose]
options:
-h, --help show this help message and exit
--verbose Enable more verbose output
Workflow
The following figure shows the overall workflow when using clinvar-this.
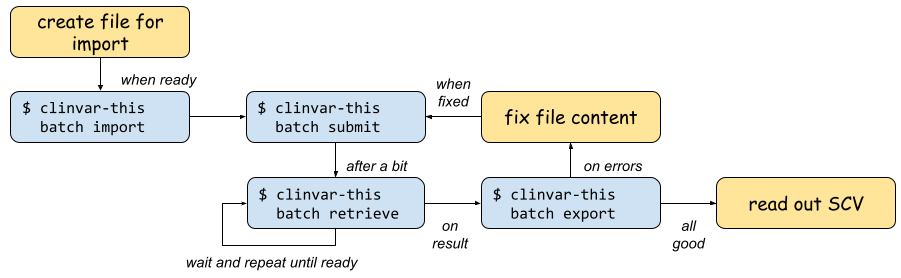
You start out by creating a file for import.
You then import the data into a local repository batch with
clinvar-this batch import.You can then post the data to the ClinVar API with
clinvar-this batch submit.The server will process your data in the background. You can query the current result with
clinvar-this batch retrieve. If this does not return yet, try again.Otherwise, you can export the current state of the batch with
clinvar-this batch exportto a TSV file.When there are errors, fix the variants to be submitted and re-submit with
clinvar-this batch submit.If everything runs to your liking, read out your ClinVar SCV identifier.
Note that the NCBI ClinVar server process runs the checks in several steps. If an earlier step fails, you will not see the results of later checks. Also, when processing runs longer this meansn that more steps succeeded so waiting longer is a good thing.
ClinVar Terminology
ClinVar is an immensely useful and large resource. The most useful upstream documentation by NCBI ClinVar includes:
A key concept in ClinVar is that it is variant-centric. That is, if you observe one variant in multiple samples (for the same condition! which may be an OMIM identifier or “not reported”!) it is one ClinVar record and ClinVar consider this record from your organisation as one submission. For your submission, you obtain a so-called SCV identifier and this is what you need for publications etc. ClinVar has these nice star ratings and these do not incorporate information on how many samples you report but only whether or not you apply formal assertion criteria such as the ACMG criteria or a local formal list of criteria. By yourself, you can only generate one star by providing an assertion criteria.
ClinVar then aggregates all submissions by all organisations for a given variant in a given condition (as explained above, yes the exlamation marks were intentional) into a reference record with an RCV record. Aggregation is based on conflicting interpretations and whether the submitters applied formal criteria (cf. nudging). In the case of multiple submitters providing conflict-free interpretations and assertion criteria, two stars may be gained. Submitters that don’t provide assertion criteria are overruled by those who do.
ClinVar will further aggreate all reference records into a variant record with a VCV identifier. For example, for BRCA variants all of these different tumor-related disorders will be thrown together.
Note that nothing in the above talked about multiple variants. You can submit them together via the NCBI submission API but each of these variants will be one NCBI ClinVar submission from your organisation. The software package clinvar-this (completely indepent of NCBI and developed on the other side of the atlantic) calls a list of variants to be submitted (as one submission each) a batch.
All of this hopefully leaves you less confused as before.
Configuration
The configuration will be stored in ~/.config/clinvar-this/config.toml in TOML format.
The file can have multiple sections, each one configuring a profile.
You should probably configure a default profile.
You can set values using clinvar-this config set NAME VALUE and read values with clinvar-this config get NAME.
A minimal configuration file looks as follows:
[default]
auth_token = "01234567890abcdefghijklm0987654321"
Before you can use clinvar-this for the first time, you have to configure the API token to use with the ClinVar submission API.
$ clinvar-this set auth_token YOURTOKENHERE
Note that configuration values will be shown in full when using varfish-cli config get/set.
Subsequently when using the tool for API submission, it will only show the first 5 characters of the secret key.
This allows to determine whether the right key is used but the value is safe enough to go to local log files etc.
However, you should still ensure to take appropriate care when exposing these 5 first characters as applicable.
Local Repository
clinvar-this creates a local repository of data in ~/.local/share/clinvar-this/$profile where $profile is the name of the profile that you use.
Below this path, you will find one directory for each submission that you manage.
Each such submission directory contains the following files:
payload.$timestamp.jsonThe payload (to be) sent to ClinVar API server at the given timestamp. The lexicographically largest file is the latest one.
On each import, a new payload file will be created. If a previous one exists, the latest one will be merged with the new to-be-imported data. Also, when the NCBI server returns SCV identifiers on success or failures, this information will be stored in a new payload file.
You can safely manipulate these JSON files but that will require some knowledge about the ClinVar API format. However, it really is not hard and with some ClinVar/bioinformatics experience, you will be able to figure it out.
submission-response.$timestamp.jsonThe response returned by the ClinVar API server returned at the given timestamp on submission. There is no direct correlation between the payload and submission response files at the moment, but you probably can figure it out based on the timestamp.
retrieve-response.$timestamp.jsonThe response returned by the ClinVar API server when calling
batch retrieve BATCH.
You can specify submission names when creating them (which is recommended). Otherwise, a name will be created for you based on the current date and time.
File Formats
See the dedicated section File Formats.
Submission Types
The following is written with the native TSV file format in mind. This translates to the other known file formats in the case that the File Formats describes the relevant columns/information.
Novel Submissions
If your sample sheet does not have a clinvar_accession column or it is empty for your variant, the variant will be submitted as novel.
ClinVar will check whether your organisation has submitted this variant before for the same condition (OMIM code or “not provided”) and report back errors if one such record exists.
clinvar-this will write the SCV from the clinvar processing results to its local repository. On re-submission of the batch after processing and result retrieval, the variant will be submitted as an update.
Submission Updates
If you provided a clinvar_accession then clinvar-this will submit an update.
Such a variant must already exist from your organisation for the given condition (again, OMIM codde or “not provided”).
Record Removals
Removals have to take another path. You have to create a removal TSV file as documented in File Formats (you only have to provide the SCV identifier to delete for and a free-text comment), import it into a new clinvar-this batch and submit it.
Note that ClinVar refers to this as “deletion” but we refer as this to “removal” to have one less term collision to the meaning of “sequence deletion”.